scatteR: Generating instance space based on scagnostics
Janith Wanniarachchi
BSc. Statistics (Hons.)
University of Sri Jayewardenepura
Sri Lanka
Session 36 (Synthetic Data and Text Analysis)
at useR! conference 2022
on the 23rd of June
What exactly are these scagnostics?

The late Leland Wilkinson developed graph theory based scagnostics that quantifies the features of a scatterplot into measurements ranging from
0 to 1

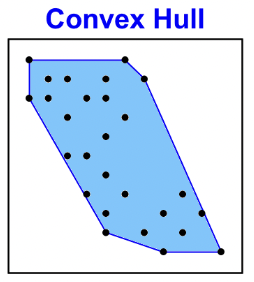
An example scatterplot
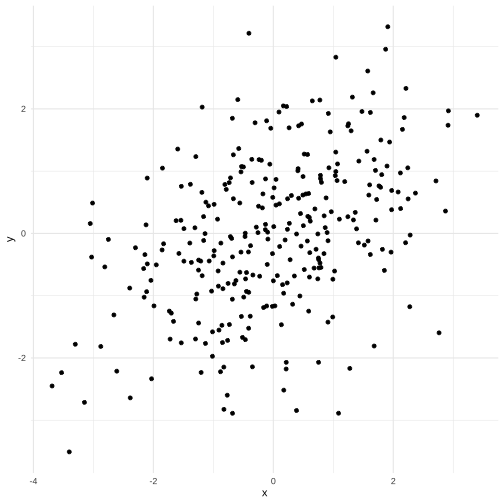
The scagnostics

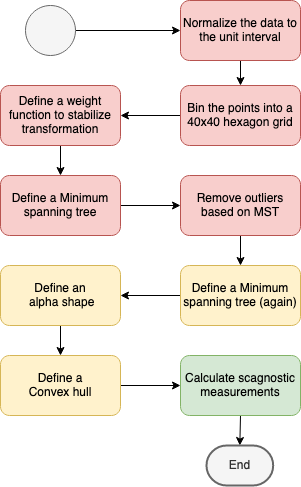
How does scagnostics work?
So how did I end up here?
As part of my research I needed a way to give marks for different features of a bivariate dataset.
That is where Scagnostics came into play.

As the next step I wanted to generate bivariate datasets which would have scagnostic values that I would expect.
Basically an inverse scagnostics!

Are there any existing solutions to this?
Are there any existing solutions to this?
Surprisingly there aren't any.

Are there any existing solutions to this?
Surprisingly there aren't any.

That's where I found my sweet
research gap
How do we generate data from this?
How do we generate data from this?
Earlier given N number of (X,Y) coordinate pairs
we got a 9×1 vector of scagnostic values
How do we generate data from this?
Earlier given N number of (X,Y) coordinate pairs
we got a 9×1 vector of scagnostic values
Now when we give a 9×1 vector of scagnostic values
we need to get N number of (X,Y) data points!
So we have to reverse a function right?
Sounds pretty simple. We can reverse a function like this
if f(x)=4x+3
So we have to reverse a function right?
Sounds pretty simple. We can reverse a function like this
if f(x)=4x+3
then
f−1(x)=x−34
But how do we actually calculate scagnostics?
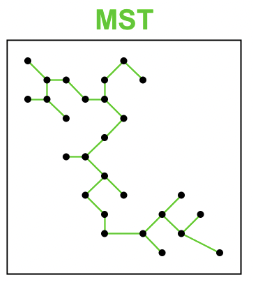
Outlying
coutlying=length(Toutliers)length(T)
Here length(Toutliers) is the total length of edges adjacent to outlying points and length(T) is the total length of edges of the final minimum spanning tree
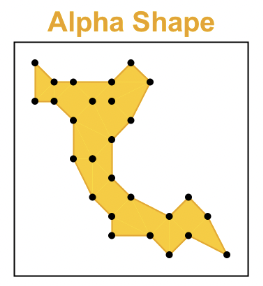
Convex
cconvex=w×area(A)area(H)
The convexity measure is based on the ratio of the area of the alpha hull and the area of the convex hull.
Monotonic
cmonotonic=r2Spearman
This is the only measurement not based on the geometrical graphs.
Skinny
cskinny=1−√4πarea(A)perimeter(A)
The ratio of perimeter to area of a polygon measures, roughly, how skinny it is.
But these equations aren't one to one functions!
Unlike in f(x)=4x+3
where for every f(x) value
we have a unique distinct x value,
But these equations aren't one to one functions!
Unlike in f(x)=4x+3
where for every f(x) value
we have a unique distinct x value,
Here there might be multiple datasets that might satisfy
all nine of a given specified scagnostic values.
This is getting out of hand!

Inspiration can come at the hungriest moments
The idea actually came to me while having desert

Inspiration can come at the hungriest moments
The idea actually came to me while having desert

Why don't I first sprinkle a little bit of data points on a 2D plot,
making sure that they land in the right places
and add on top of those sprinkles (data points)
keep on adding more sprinkles
so that the final set of sprinkles (data points) looks good!
But how do we arrange these points in the most optimal manner?
Given a set of N number of (X,Y) data points and a 9×1 vector of expected scagnostic values, we need to minimize the distance between the scagnostic vector of the current dataset and the expected scagnostic measurement.
But how do we arrange these points in the most optimal manner?
Given a set of N number of (X,Y) data points and a 9×1 vector of expected scagnostic values, we need to minimize the distance between the scagnostic vector of the current dataset and the expected scagnostic measurement.
Let's define the loss function as,
L([X–– Y––])=1kk∑i=1|si([Di−1[X–– Y––]])−m0i|
where Dt=[Dt−1,X––,Y––], i∈{Outlying,Skewed,…,Monotonic} , si([Di−1;[X–– Y––]]) and m0i is the ith calculated and expected scagnostic measurement respectively.
But how do we arrange these points in the most optimal manner?
Given a set of N number of (X,Y) data points and a 9×1 vector of expected scagnostic values, we need to minimize the distance between the scagnostic vector of the current dataset and the expected scagnostic measurement.
Let's define the loss function as,
L([X–– Y––])=1kk∑i=1|si([Di−1[X–– Y––]])−m0i|
where Dt=[Dt−1,X––,Y––], i∈{Outlying,Skewed,…,Monotonic} , si([Di−1;[X–– Y––]]) and m0i is the ith calculated and expected scagnostic measurement respectively.
so now we need to find a optimizer for the 2N parameters of x1,x2,...,xN,y1,y2,...,yN
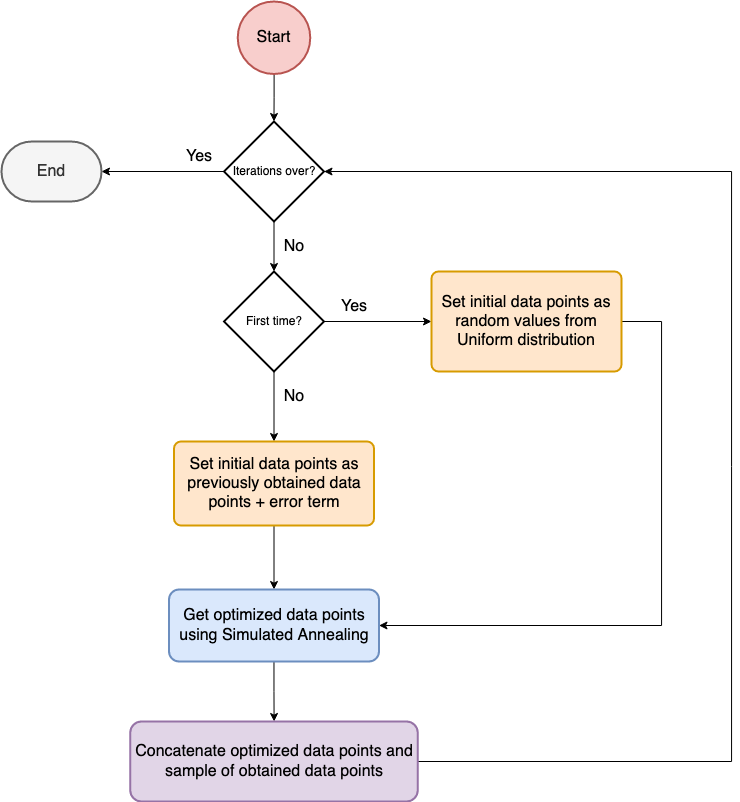
Simulated Annealing

The name of the algorithm comes from annealing in material sciences, a technique involving heating and controlled cooling of a material to alter its physical properties.
The algorithm works by setting an initial temperature value and decreasing the temperature gradually towards zero.
As the temperature is decreased the algorithm becomes greedier in selecting the optimal solution.
In each time step, the algorithm selects a solution closer to the current solution and would accept the new solution based on the quality of the solution and the temperature dependent acceptance probabilities.

The algorithm
A simple example
library(scatteR)library(tidyverse)df <- scatteR(measurements = c("Monotonic" = 0.9),n_points = 200,error_var = 9)qplot(data=df,x=x,y=y)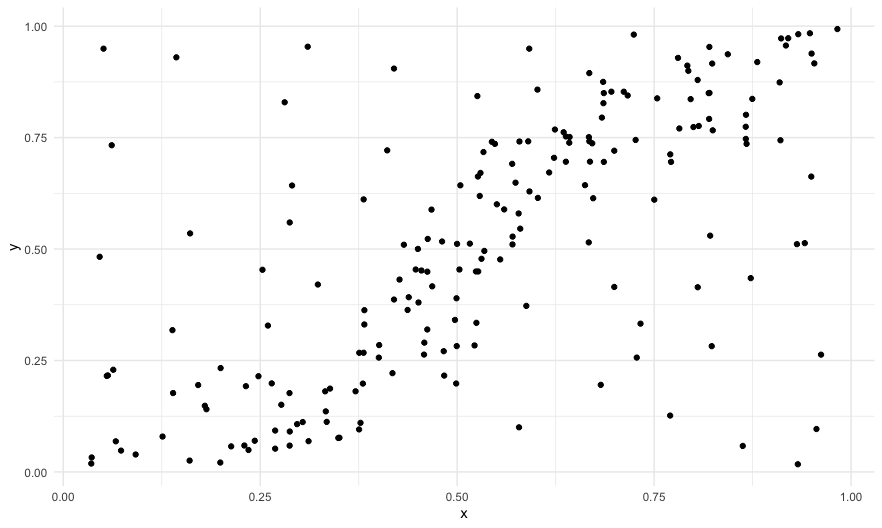
Behind the scenes,
The simulated annealing component is achieved through the GenSA package.
Y. Xiang, et al (2013). Generalized Simulated Annealing for Efficient Global Optimization: the GenSA Package for R.
The scatteR() function
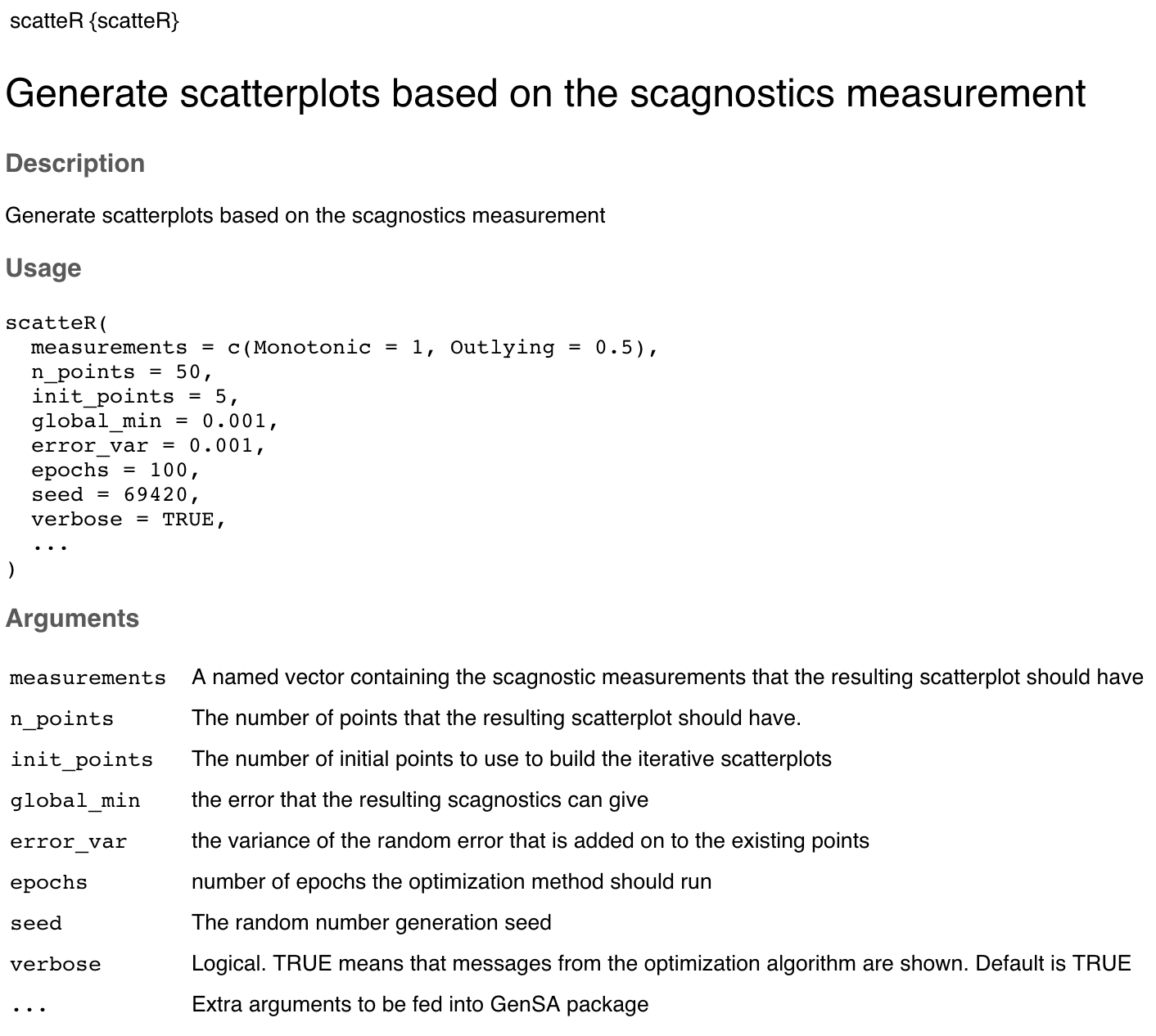
Here I will be showcasing the documentation and the arguments that are available and what each of those arguments mean.
Performance results
Type wise error
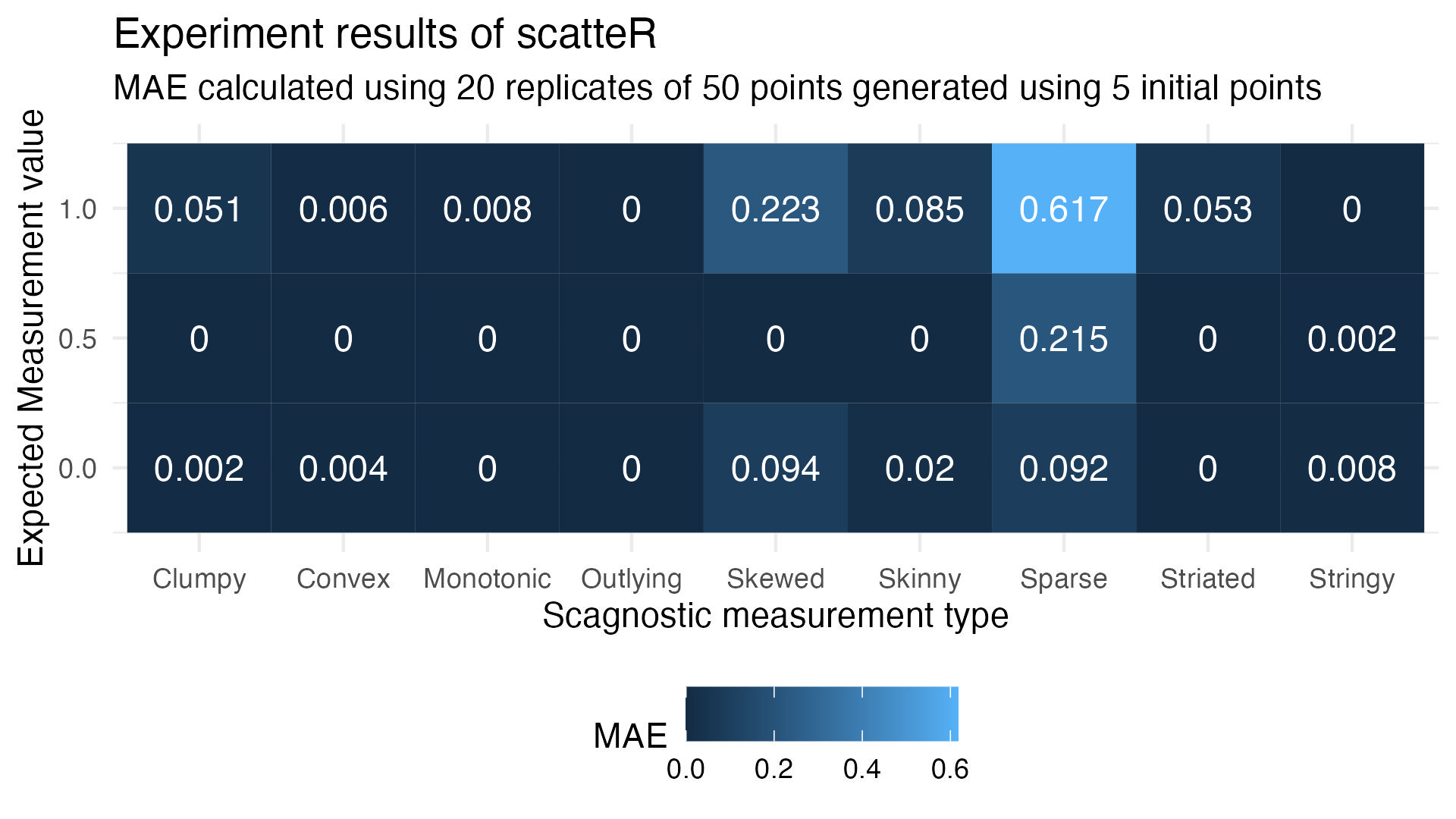
Is there a special recipe for the hyperparameters
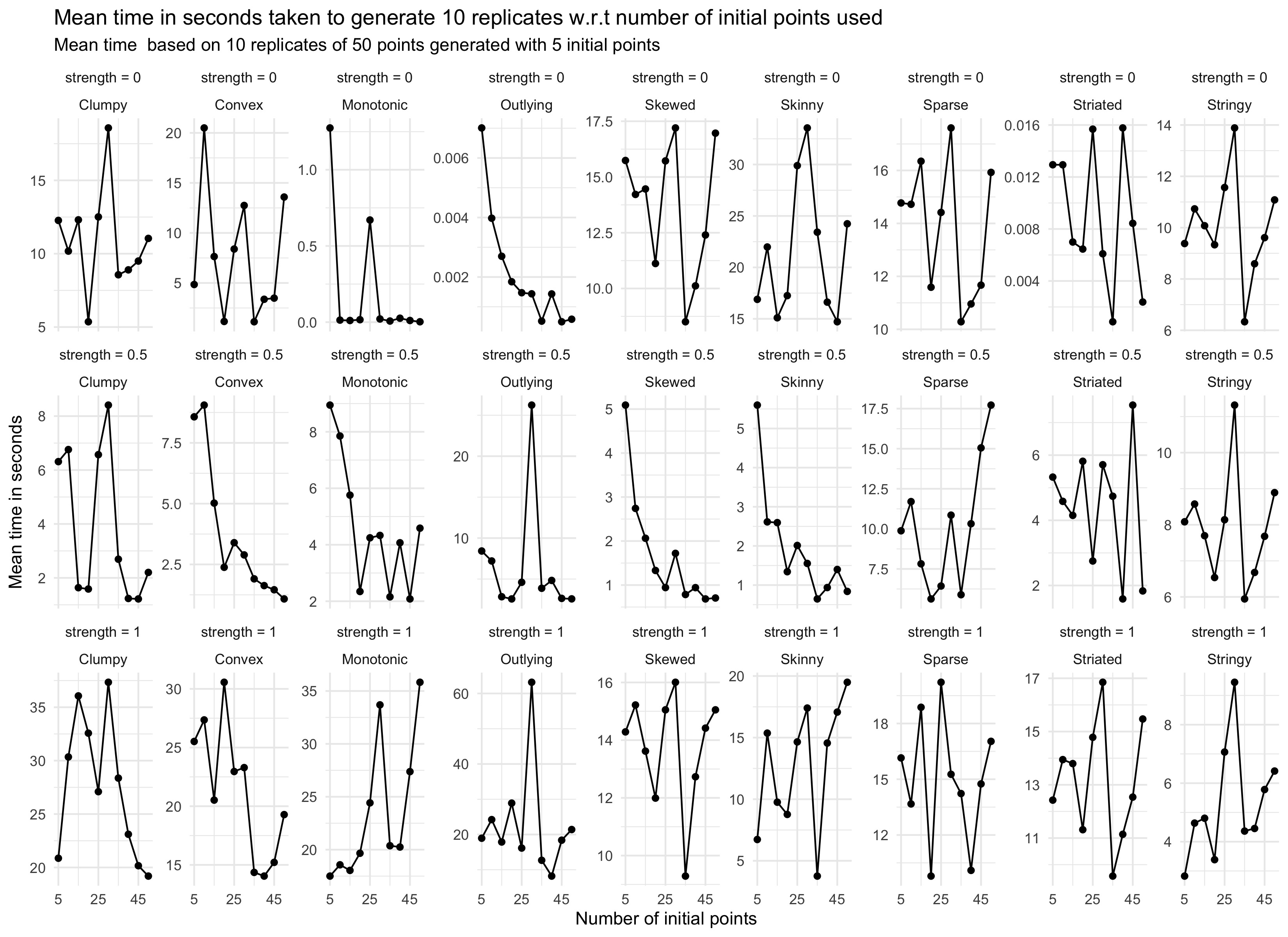
Here I will be talking about the effects of changing different parameters.
How much time does it take?
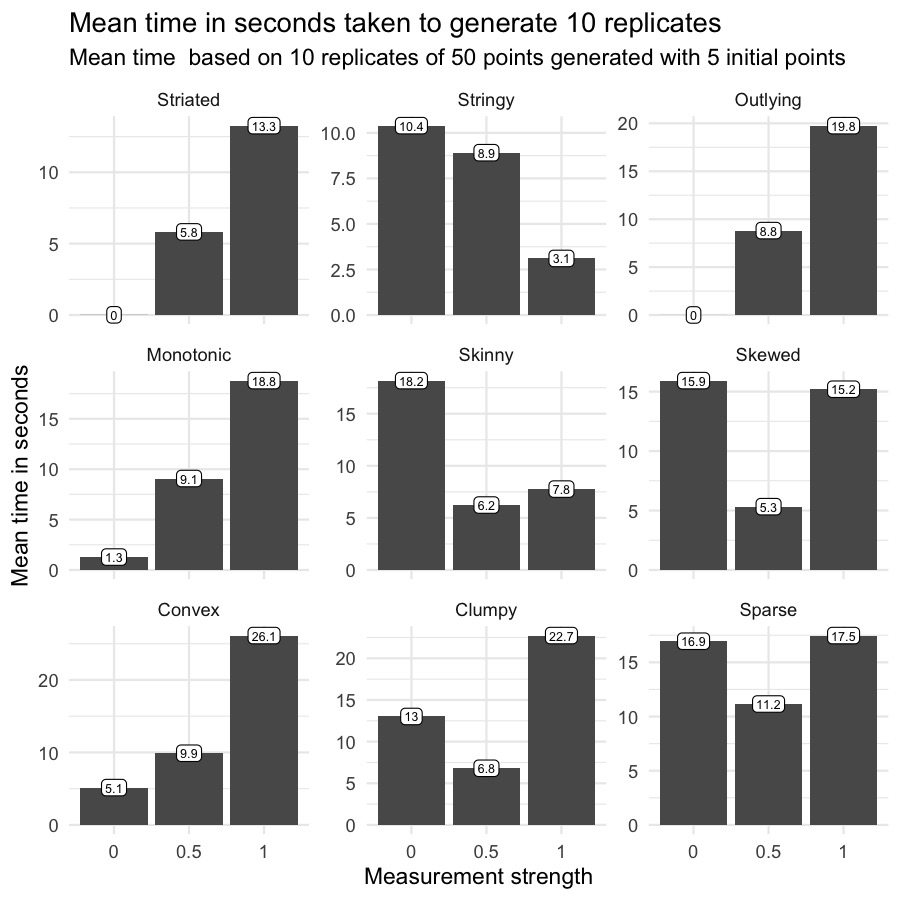
Here I will be talking about the time complexity of scatteR and the ways to speed the generation process
In summary, what can scatteR do for you?
As a teacher,
- Generate dummy data for the students to try out new statistical methods
- Introduce students to the concept of scagnostics
In summary, what can scatteR do for you?
As a teacher,
- Generate dummy data for the students to try out new statistical methods
- Introduce students to the concept of scagnostics
As a data scientist,
- Synthesize small scale numerical datasets for test purposes
- Generate dummy data to try out new data science methods
In summary, what can scatteR do for you?
As a teacher,
- Generate dummy data for the students to try out new statistical methods
- Introduce students to the concept of scagnostics
As a data scientist,
- Synthesize small scale numerical datasets for test purposes
- Generate dummy data to try out new data science methods
As an everyday R user,
- Generating data for an interesting generative art made with R
- Generate a quick sample of data to test out a new package that you installed
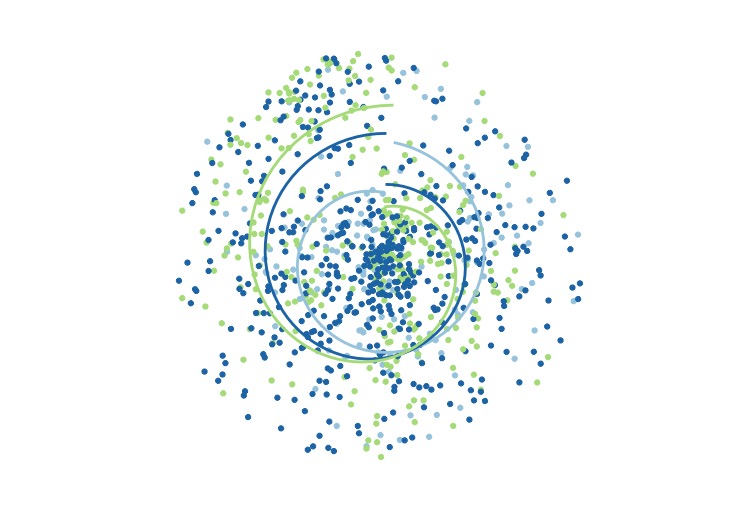
A generative art based on the bivariate numeric relationships of the palmerpenguins dataset
Where to from here?
Better optimization methods
Parallelized implementations
Replacing the relevant R code to C++ code
and many more so stay tuned!

Thank you for listening!
Thank you to useR! 2022 and sponsors

For awarding me with the diversity scholarship that gave me the financial strength to speak before you
Have any follow up questions?
Email: janithcwanni@gmail.com
Twitter: @janithcwanni
Github: @janithwanni
Linkedin: Janith Wanniarachchi
Try scatteR at https://github.com/janithwanni/scatteR
Slides available at: https://scatter-use-r-2022.netlify.app/
Created with xaringan and xaringan themer
Acknowledgements
The following content were created by the respective creators and not my work.
- Image of Ice cream sprinkles (Slide #17): Photo by David Calavera on Unsplash
- Image of Annealing (Slide #19): http://www.turingfinance.com/simulated-annealing-for-portfolio-optimization/
- Image of Caterpie staring at moon made by All0412 on deviantart (Slide #24) https://www.deviantart.com/all0412/art/Caterpie-353761155

